Virtual screening
SAMPL4
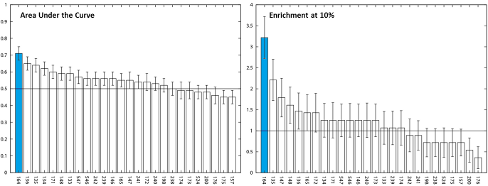
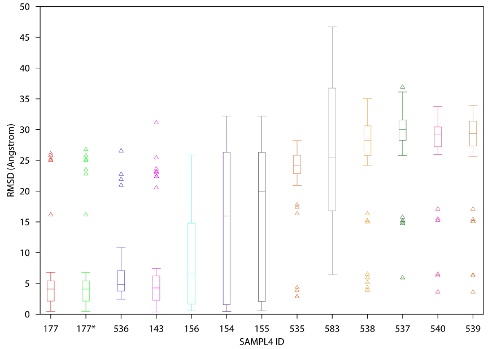
The SAMPL (Statistical Assessment of the Modeling of Proteins and Ligands) challenges provide an ideal opportunity for unbiased evaluation and comparison of different approaches used in computational drug design. During the fourth round of this SAMPL challenge, we participated in the virtual screening and binding pose prediction on inhibitors targeting the HIV-1 integrase enzyme. For virtual screening, we used well known and widely used in silico methods combined with personal in cerebro insights and experience. Regular docking only performed slightly better than random selection, but the performance was significantly improved upon incorporation of additional filters based on pharmacophore queries and electrostatic similarities. The best performance was achieved when logical selection was added. For the pose prediction, we utilized a similar consensus approach that amalgamated the results of the Glide-XP docking with structural knowledge and rescoring. The pose prediction results revealed that docking displayed reasonable performance in predicting the binding poses. However, prediction performance can be improved utilizing scientific experience and rescoring approaches. In both the virtual screening and pose prediction challenges, the top performance (ranked No. 1) was achieved by our approaches (ID 164 in virtual screening and ID 177 in pose prediction as shown in figures below). This work has been published in JCAMD
(publication).