Virtual screening
CSARdock2014
Application of Shape Similarity in Pose Selection and Virtual Screening in CSARdock 2014 Exercise
To evaluate the applicability of shape similarity in docking based pose selection and virtual screening, we participated in the CSARdock2014 benchmark exercise for identifying the correct docking pose of inhibitors targeting factor XA, spleen tyrosine kinase and tRNA methyltransferase. This exercise provides a valuable opportunity for researchers to test their docking programs, methods, and protocols in a blind testing environment. In the CSARdock2014 benchmark exercise, we have implemented an approach that uses ligand 3D shape similarity to facilitate docking based pose selection and virtual screening. We showed here that ligand 3D shape similarity between bound poses could be used to identify the native like pose from an ensemble of docking generated poses. Our method correctly identified the native pose as the top-ranking pose for 73% of test cases in a blind testing environment. Moreover, the pose selection results also revealed an excellent correlation between ligand 3D shape similarity scores and RMSD to X-ray crystal structure ligand. In the virtual screening exercise, the average RMSD for our pose prediction was found to be 1.02 Å and it was one of the top performances achieved in CSARdock2014 benchmark exercise. Furthermore, the inclusion of shape similarity improved virtual screening performance of docking based scoring and ranking. The coefficient of determination (r2) between experimental activities and docking scores for 276 spleen tyrosine kinase inhibitors was found to be 0.365 but reached 0.614 when the ligand 3D shape similarity was included. This work has been published in JCIM (publication).
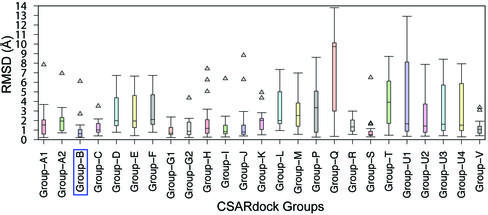
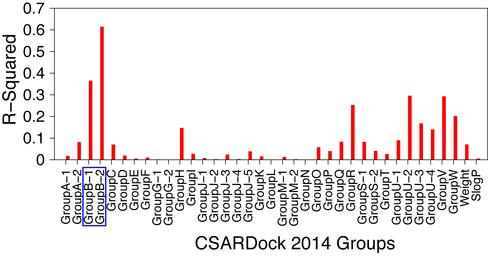
To evaluate the applicability of shape similarity in docking based pose selection and virtual screening, we participated in the CSARdock2014 benchmark exercise for identifying the correct docking pose of inhibitors targeting factor XA, spleen tyrosine kinase and tRNA methyltransferase. This exercise provides a valuable opportunity for researchers to test their docking programs, methods, and protocols in a blind testing environment. In the CSARdock2014 benchmark exercise, we have implemented an approach that uses ligand 3D shape similarity to facilitate docking based pose selection and virtual screening. We showed here that ligand 3D shape similarity between bound poses could be used to identify the native like pose from an ensemble of docking generated poses. Our method correctly identified the native pose as the top-ranking pose for 73% of test cases in a blind testing environment. Moreover, the pose selection results also revealed an excellent correlation between ligand 3D shape similarity scores and RMSD to X-ray crystal structure ligand. In the virtual screening exercise, the average RMSD for our pose prediction was found to be 1.02 Å and it was one of the top performances achieved in CSARdock2014 benchmark exercise. Furthermore, the inclusion of shape similarity improved virtual screening performance of docking based scoring and ranking. The coefficient of determination (r2) between experimental activities and docking scores for 276 spleen tyrosine kinase inhibitors was found to be 0.365 but reached 0.614 when the ligand 3D shape similarity was included. This work has been published in JCIM (publication).

