Virtual screening
D3R2015
Prospective Evaluation of Shape Similarity Based Pose Prediction Method in D3R Grand Challenge 2015
Evaluation of ligand 3D shape similarity is one of the commonly used approaches to identify ligands similar to one or more known active compounds from a library of small molecules. Apart from using ligand shape similarity as a virtual screening tool, its role in pose prediction and pose scoring has also been reported. We have recently developed a method that utilizes ligand 3D shape similarity with known crystallographic ligands to predict binding poses of query ligands. Here, we report the prospective evaluation of our pose prediction method through the participation in D3R Grand Challenge 2015. Our pose prediction method was used to predict binding poses of HSP90 and MAP4K4 ligands and it was able to predict the pose within 2 Å RMSD either as the top pose or among the best of five poses in a majority of cases. Specifically for HSP90 protein, a median RMSD of 0.73 and 0.68 Å was obtained for the top and the best of five predictions respectively. For MAP4K4 target, although the median RMSD for our top prediction was only 2.87 Å but the median RMSD of 1.67 Å for the best of five predictions was well within the limit for successful prediction. Furthermore, the performance of our pose prediction method for HSP90 and MAP4K4 ligands was always among the top five groups. Particularly, for MAP4K4 protein our pose prediction method was ranked number one both in terms of mean and median RMSD when the best of five predictions were considered. Overall, our D3R Grand Challenge 2015 results demonstrated that ligand 3D shape similarity with the crystal ligand is sufficient to predict binding poses of new ligands with acceptable accuracy. This work has been published in JCAMD (publication).
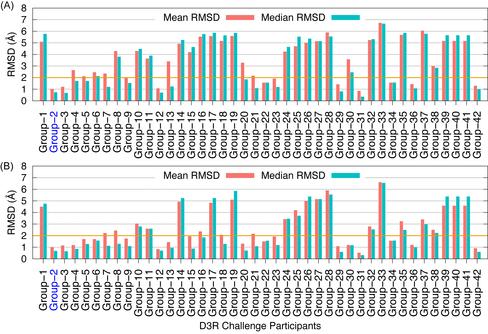
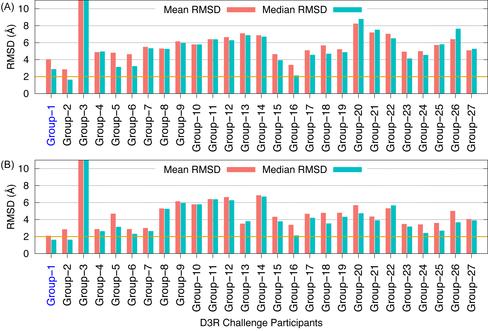
Evaluation of ligand 3D shape similarity is one of the commonly used approaches to identify ligands similar to one or more known active compounds from a library of small molecules. Apart from using ligand shape similarity as a virtual screening tool, its role in pose prediction and pose scoring has also been reported. We have recently developed a method that utilizes ligand 3D shape similarity with known crystallographic ligands to predict binding poses of query ligands. Here, we report the prospective evaluation of our pose prediction method through the participation in D3R Grand Challenge 2015. Our pose prediction method was used to predict binding poses of HSP90 and MAP4K4 ligands and it was able to predict the pose within 2 Å RMSD either as the top pose or among the best of five poses in a majority of cases. Specifically for HSP90 protein, a median RMSD of 0.73 and 0.68 Å was obtained for the top and the best of five predictions respectively. For MAP4K4 target, although the median RMSD for our top prediction was only 2.87 Å but the median RMSD of 1.67 Å for the best of five predictions was well within the limit for successful prediction. Furthermore, the performance of our pose prediction method for HSP90 and MAP4K4 ligands was always among the top five groups. Particularly, for MAP4K4 protein our pose prediction method was ranked number one both in terms of mean and median RMSD when the best of five predictions were considered. Overall, our D3R Grand Challenge 2015 results demonstrated that ligand 3D shape similarity with the crystal ligand is sufficient to predict binding poses of new ligands with acceptable accuracy. This work has been published in JCAMD (publication).

