Research Topics
Development and application of analytical methods for structural details on biological molecules
Development of quantitative analysis of biomolecules
Identification and characterization of RNA by mass spectrometry
Development and application of analytical methods for structural details on biological molecules (Dohmae, Suzuki, Watanabe, Kwak,Kubota)
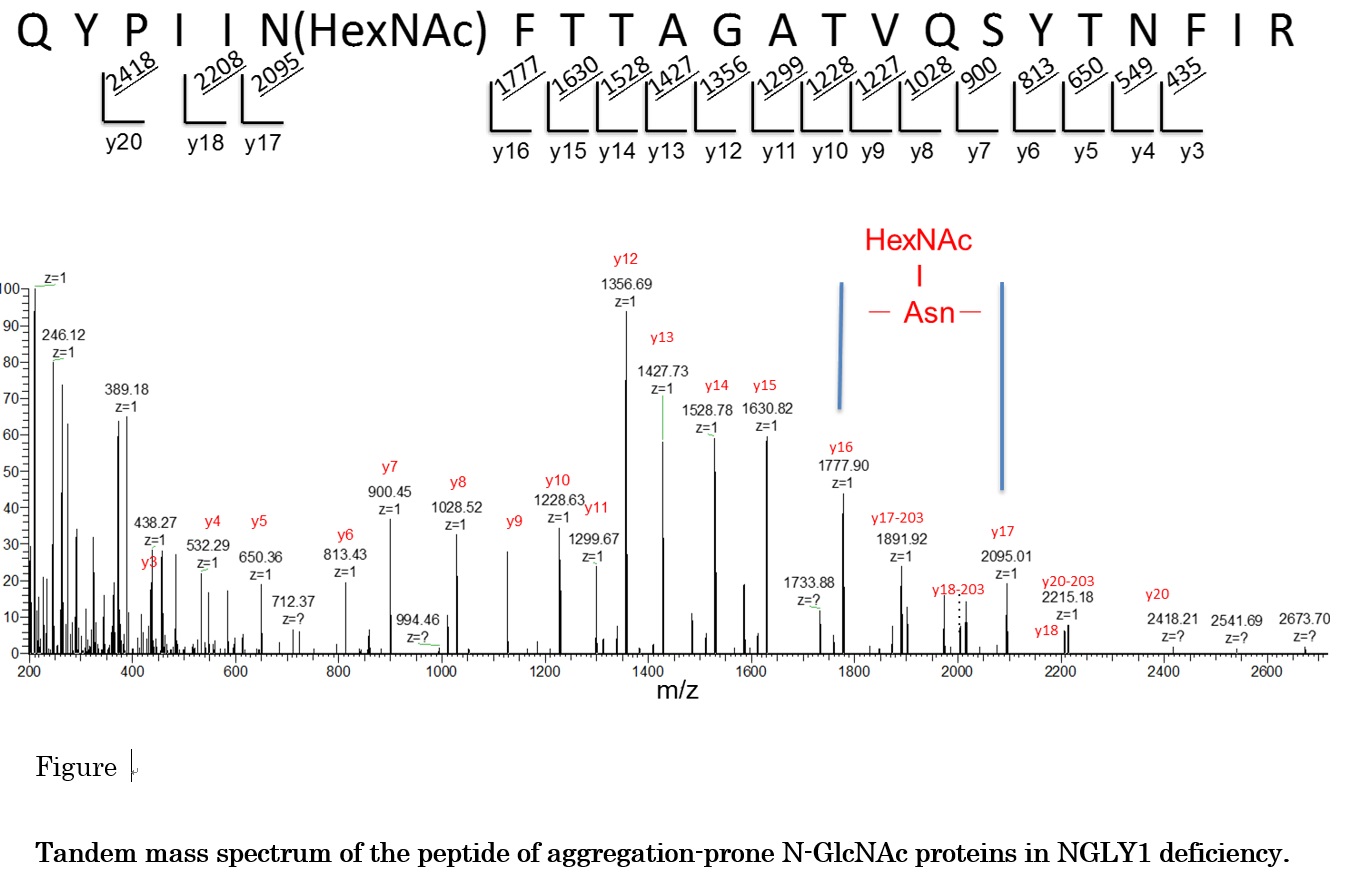
Genetic code is transcripted into RNA followed by translation to proteins. Many proteins capture appropriate functions and suffer regulations by post-translational modifications (PTMs). Direct analysis of the modifications of amino acid residues and their positions within a protein sequence is still required in post genome sequence era. Furthermore, binding site of a low molecular reagent to a protein is the most important information for an approach using chemical biology with bio-probe. In epigenome , PTMs of histone or non-histone protein are focused due to control of gene expression. So, we have been developing the characterization methods of PTMs and have been applying it for biological problems. Since the 2012, we have gone in a research project, the research on the innovative development and the practical application of new drugs for Hepatitis B. In this year, we start to search a CDE (cccDNA Eliminator) compound binding site of target protein using photo-affinity labeled methods.We also joined P-DIRECT (Project for Development of Innovative Research on Cancer Therapeutics) with Graduate School of Science, the University of Tokyo and the University of Chicago and we determined methylation site of the PTEN protein for cancer related methyl-transferase SMYD2. And in a collaboration with Dr. T. Suzuki, Glycometabolome Team/RIKEN GRC, we found that endo-β-N-acetylglucosaminidase (ENGase) was partially stripping sugars directly off the proteins in mouse cells without N-glycanase activity. The unconventional deglycosylation reaction was found to be catalyzed by the cytosolic ENGase, generating aggregation-prone N-GlcNAc proteins and we press-released “Drug target found for rare genetic disorder (Blocking an enzyme involved in protein recycling could provide therapeutic relief for people with NGLY1 deficiency)”.
Development of quantitative analysis of biomolecules (Dohmae, Kawata, Banzai, Suzuki, Kwak)
Quantification of post-translational modifications (PTMs) of proteins is necessary for epigenetics or proteomics research. To complement qualitative analyses by mass spectroscopy such as determination of positions or kinds of PTMs, we have developed ultra-sensitive amino acid analysis in which absolute quantity of less than a hundred nanograms of proteins were successfully determined. In the project of P-DIRECT, we have applied target MS/MS to quantify in vivo protein methylation in the cells overexpressing methylation enzyme and substrate genes. We successfully quantified the histone variant TH2B that is abundant in the testis and solved the function of TH2A and B during spermiogenesis (joint-research with Dr. Dr. Shunsuke Ishii, Riken Tsukuba Center)
RNA by mass spectrometry (Nakayama, Koike、Shiina)
We have developed a database search engine, Ariadne that enable the identification of an RNA using tandem mass spectrometry data of its RNase digested nucleotides by searching a DNA/RNA database such as genome DNA sequence. We are expanding the capability of our RNA analytical platform to identify small RNA such as miRNA and its modifications directly. We have reported an analytical system for the direct identification of miRNAs that incorporates nanoflow liquid chromatography - high-resolution tandem mass spectrometry with a spray-assisting device that stabilizes negative nanoelectrospray ionization of RNAs and modified version of the database searching. The system has successfully identified femtomole quantities of human cellular miRNAs and their 3′-terminal variants from HeLa cells. This is the first report of a fully automated, and thus objective, tandem mass spectrometry-based analytical system that can be used to identify miRNAs. → Link to Ariadne service


