Research Topics
Development and application of analytical methods for structural details on biological molecules
Development of quantitative analysis of biomolecules
Studies on structural biology and its related technologies
Identification and characterization of RNA by mass spectrometry
1. Development and application of analytical methods for structural details on biological molecules (Dohmae, Suzuki, Masuda, Watanabe, Kwak)
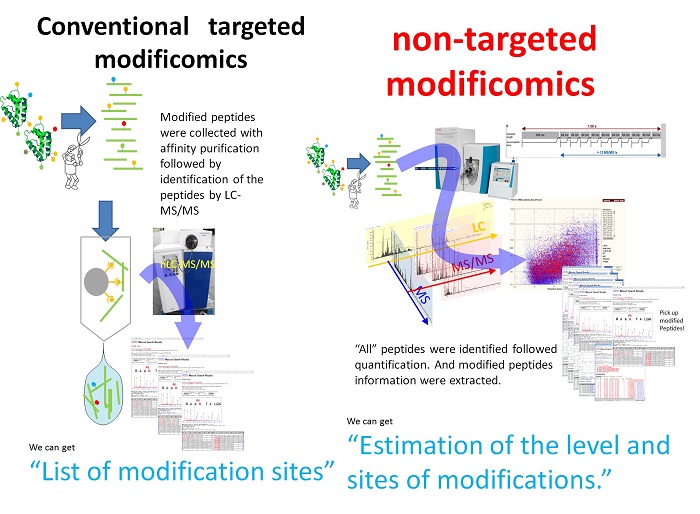
Genetic code is transcripted into RNA followed by translation to proteins. Many proteins capture appropriate functions and suffer regulations by post-translational modifications (PTMs). Direct analysis of the modifications of amino acid residues and their positions within a protein sequence is still required in post genome sequence era. Furthermore, binding site of a low molecular reagent to a protein is the most important information for an approach using chemical biology with bio-probe. In the epigenome field, PTMs of histone or non-histone protein are focused to control the gene expression. So, we have been developing the characterization methods of PTMs and have been applying it for biological problems.
We have gone in a research project, the research on the innovative development and the practical application of new drugs for hepatitis B, and started global analysis of PTMs. In this year, we have developed the non-targeted modificomics that extracted modified peptides information from identified all peptides in shotgun proteomics. This method differs from the conventional targeted modificomics that is pre-purification with affinity column (i.e. IMAC etc.) followed by shotgun proteomics. We observed the modification level that was a ratio of the modified peptides versus normal one of the protein in comparative cells, which we could not observe in targeted modificomics.
We also joined P-DIRECT (Project for Development of Innovative Research on Cancer Therapeutics) with Graduate School of Science, the University of Tokyo and the University of Chicago and we determined methylation site of the target protein for cancer related methyl-transferases. And in collaboration with Graduate School of Science, the University of Tokyo, we examined the binding materials with YidC that is the inner membrane protein of Escherichia coli involved in the integration of membrane proteins with SecYE and that can detect unknown electron cloud in X-ray analysis of YidC crystal. We press released "Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli." collaborating with Institute for Virus Research Kyoto University.
Development of quantitative analysis of biomolecules (Masuda, Suzuki, Dohmae, Kawata, Banzai)
Quantification of post-translational modifications (PTMs) of proteins is necessary for epigenetics or proteomics research. To complement qualitative analyses by mass spectroscopy such as determination of positions or kinds of PTMs, we have developed ultra-sensitive amino acid analysis in which absolute quantity of less than a hundred nanograms of proteins were successfully determined. To quantify a small amount of PTM-amino acids in the presence of a large amount of other normal amino acids, we developed 2D chromatography in which amino acids of a hydrolyzed sample are separated by a graphite column before AAA using reversed-phase chromatography. We have developed a hydrolysis method of proteins using solid acid catalyst. In the current year, we examined hydrolysis efficiency of a new catalyst that has highly heat resistance. We and Dr. Akeo Shinkai of Riken Spring-8 center determined structure and function of Cmr complex which contributes to bacterial antiviral defense. We also successfully quantified tryptophan (joint-research with Dr. Tadashi Suzuki, Systems Glycobiology Research Group, Riken), and methylated arginine and/or methylated lysine (joint-research with Dr. Ryuji Hamamoto of Univ. Tokyo).
Studies on structural biology and its related technologies(Miyatake)
SPring-8 is the most excellent synchrotron facility in the world, but researchers on the off-site often face difficulties to collect data at the maximum efficiency in the limited beam-times. Thus our team and the division of synchrotron radiation instrumentation (SPring-8 center) together built a mail-in data collection system in Wako campus area for remote accumulation of the diffraction data at SPring-8. On the other hand, crystallization of target proteins is one of the most difficult processes for the recent X-ray crystallography, so that automated device for macromolecular crystallization is strongly desired. In this point of view, we have developed a novel device for protein crystallization based on dynamic light scattering in corporation with a company (HM: in collaboration with YMC Co., Ltd.).
RNA by mass spectrometry (Nakayama)
We have developed a database search engine, Ariadne that enable the identification of an RNA using tandem mass spectrometry data of its RNase digested nucleotides by searching a DNA/RNA database such as genome DNA sequence. We have expanded the capability of Ariadne to identify miRNAs and their modifications directly. → Link to Ariadne service


