Prediction and design
PIZZA
Computational design of a self-assembling symmetrical β-propeller protein
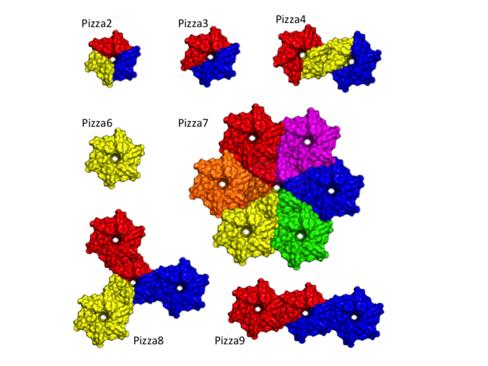
The modular structure of many protein families such as β-propeller proteins strongly implies that duplication played an important role in their evolution, leading to highly symmetrical intermediate forms. Previous attempts to create perfectly symmetrical propeller proteins have failed however. We have therefore developed a new and rapid computational approach to design such proteins. As a test case, we have created a 6-fold symmetrical β-propeller protein (named PIZZA6), and experimentally validated the structure using X-ray crystallography. Each blade consists of 42 residues. Proteins carrying 2 to 10 identical blades were also expressed and purified (named PIZZA2 to PIZZA10). Two or three tandem blades assemble to recreate the highly stable 6-fold symmetrical architecture, consistent with the duplication and fusion theory. The other proteins produce different mono-disperse complexes, up to 42 blades (180 kDa) in size, which self-assemble according to simple symmetry rules. Our procedure is suitable for creating nano-building blocks from different protein templates of desired symmetry. This work has been published in PNAS (publication).This work has been highlighted by RIKEN Research published on Dec. 12, 2014. Perfect propeller proteins designed by computer
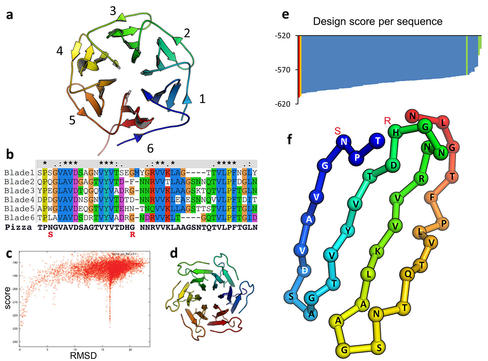
Our design process is illustrated in the figure below. Please see our paper for details.
Our designed proteins have a strong preference for the 6-bladed structure. Each complex has a size determined by the lowest common multiple (LCM) of six and the number of blades per polypeptide chain. The crystal structures of Pizza2, Pizza3 and Pizza6 are shown in the figure below. The proposed model of Pizza4, Pizza7, Pizza8 and Pizza9 are also shown.