| JAPANESE | ENGLISH |
Image Processing Research Team, Center for Advanced Photonics, RIKEN
Research
Dr. Hideo Yokota
RIKEN Image Processing Research Team: Team Leader
Research Subjects
- Algorithms and computational methods for image processing, pattern recognition, computer graphics, and computer aided design.
- Image-based software and hardware systems for biology and material science.
- Image-based systems for medical applications.
- Imaging systems for life and material sciences.
V-Cat Systems: Volume Segmentation Software
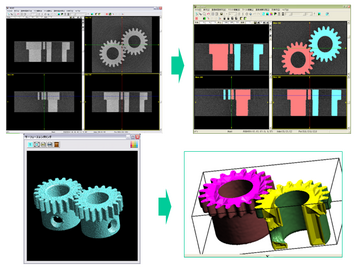
V-Cat Example
The role of V-Cat system is to reconstruct the real-world objects in the computer. There are no design schematics for the real-world objects. Even if there is a design schematic as the industrial manufacture, the precision and error problems exist in the convectional CAD/CAM/CAE process. It is important to obtain the data which can be considered as a design schematic of the real-world object. The V-Cat system plays a role of the VCAD world entrance.
- V-Cat (Standard Version)
- Academic Version
- Linux-Cluster Version
- V-Cat 4D Version
- VCAT5 (Japanese Only)
VCAT System Process
- 1. Obtain the sectional images in order to generate the volume data.
- 2. Visualize the sectional images: V-Cat employs the sectional views and perspective view of the volume.
- 3. Extract the target object (region): the volume is segmented according to the interests of the user by adding mask IDs to the volume. The basic input interface is the mouse or pen tablet to segment the volume by manually. Moreover the discriminant thresholding and a region growing methods are implemented to recognize the target objects automatically. The volume segmentation is not only for generating VCAD data but also useful for various mathematical analysis such as volume and area calculations of the segments. V-Cat already employed in the industrial and natural science applications such as the structured analysis of industrial real data consists of a lot of poles and the distribution ratio analysis of organelles in the living cell.
- 4. Generate the VCAD data: the iso-surfaces approximated by the triangle meshes are extracted according to the segmented volume. The vobj file format (a subset of the obj format) is supported. The newest version includes the function to generate the hexahedral volumetric mesh of the segmented volume. The generated volumetric mesh can be employed for the physical simulations.
Acquisition Technology for Bio-Material Internal Structure

3D-ISM (soft tissues and bones)
We have been working on various systems of 3D-ISM (Three Dimensional Internal Structure Microscope) which is a destructive testing system for bio-materials. Our 3D-ISM is able to observe micro-meter resolution with centimeter order regions by slicing the object into sectional images. For example, a single cell of a mouse corresponds to a pixel for high-resolution setting. It is easy to construct volumetric (3D) images, because color and its corresponding coordinates of the target for measurement are obtained in computer via our system. Our system is useful for many bio-related applications such as shape data for physical and biochemical simulations, digital atlas, and gene expression analysis.
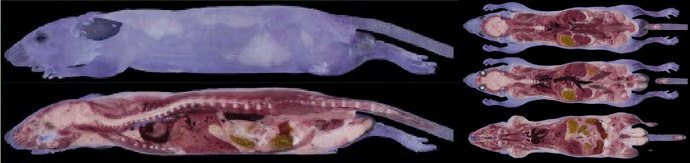
3D color image of a mouse.
Acquisition Technology for Material Internal Structure

3D Hard Tissue/Material Internal Structure Microscope

3D Internal Element Analysis
CT and MRI which have been widely used in medical science generate 3D volume information by scanning 2D sectional images. In our approach, a test material is actually sliced to construct 3D models. Soft materials such as biogenic tissues can be sliced by using razor blades. On the other hand, the razor blades are easily broken by slicing the hard materials such as bones and metals. Therefore, we employ the high precision cutting techniques. High precision cutting is the manufacturing technology which generates the resulting surface of the metal and the plastic material precisely. Our method produces the fair surfaces whose precision is 100 nm. These fair surfaces are very suitable for the microscope observation because of their smoothness and preservation of original structures. We achieve high quality internal material acquisition results which include poles, impurities, and crystal structures of slice sections. The proposed system consists of alternative processes of high precision cutting and acquisition of the sectional images makes possible to generate the 3D volume data quickly. Also the system includes the X-Ray element analysis devices which achieves to observe 3D element distribution in the test materials.
Cloud-based Image Processing and Communications
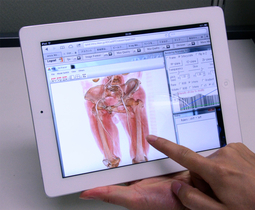
Interactive volume rendering via a tablet PC.
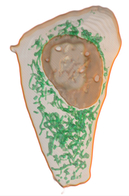

Left: HeLa cell with its mitochondria. Right: Medical CT.
Modern life-science requires to manage and analyze large-scale 3D images because of rapid advances in acquisition technologies such as confocal laser scanning microscopes, CT, and MRI. Also modeling and analysis in engineering based on real-world image data have considerable attention such as mechanical simulation and 3D rapid prototyping. Unfortunately, many existing image processing systems are not equipped with network and communication functions which are required for collaborative research and development. We have been developing a cloud-based system for image processing and communications. Our system employes standard web-browsers for a client which allows us to manage and process 3D images thought network and provides services such as sharing, filtering, visualizing, interactive segmentation, and managing processing history of 3D images. For example, volume rendering needs high-end GPUs and/or expensive PCs in general cases, but our system provides such services through network and so that the users are able to use low-end/tablet PCs as a client PC. Our system shares limited software and hardware resources via cloud-based services for efficient and effective collaboration of image-based research and development.
Application of V-Cat and Volume Segmentation to Cell Biology
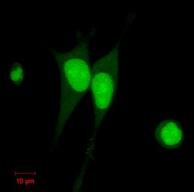
Established HeLa cell lines
expressing GFP-NLS stably.
Significance of quantitative analysis based on the microscopic data has been increasing in cell biology. Also, recent advances in the microscope and fluorescent protein technologies allow us time-lapse observation of living cells. Thus, we are developing the quantitative analysis methods which employ the V-Cat system and volume segmentation techniques with 4D observations of living cells for the investigation of cell biology. Besides these methodological contributions, our goal includes understanding for molecular mechanisms of membrane traffic and organelle formation processes based on the analysis results. Moreover, we are working on the establishment of biological experimental methods for the V-Cat system such as constructing the plasmids which label the target molecules and organelles with fluorescent proteins, and establishing the cell lines which express the specific fluorescent conjugated protein stably.
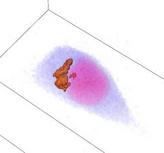

Golgi apparatus around mitosis.
Medical Image Analysis via Deep Learning Techniques
We have been working to apply deep learning technology, which has made remarkable progress in recent years, to medical image analysis, and aim to develop practical computational systems useful for medical application. Our approach is based on a popular learning method called CNN (Convolutional Neural Network) by modeling and applying its network models, learning protocols, data augmentations, and prediction frameworks, for various diseases and image/problem settings specific to medical devices.
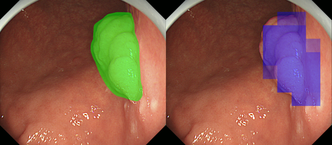
Fig. 1 Detection result (right) and training (left) images.
Early gastric cancer detection in endoscopic images: Detecting early gastric cancers using endoscopic images is difficult even for gastroenterologists because their morphological features are inconspicuous compared with colorectal or advanced gastric cancers. In this research, we proposed a CNN-based detection method of early gastric cancer in endoscopic images in collaboration with RIKEN and NCCHE (National Cancer Center Hospital East). Obtaining a large number of high quality training images of early gastric cancer required for deep learning is not realistic. Therefore, we have succeeded in automatically detecting cancer regions with high precision and efficiency by applying a data augmentation technique. Our result hopefully contributes to early detection and treatment by reducing missed gastric cancers in screening.
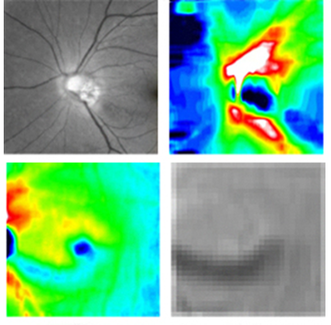
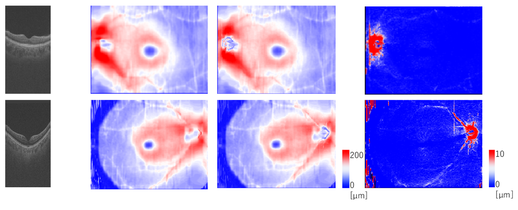
Fig.2 Multi-modal image data (glaucoma).
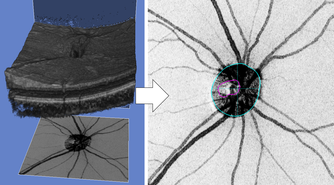
Fig.3 Quantitative ocular parameters from the observed images.
Glaucoma Detection via Multi-modal Image Information: Since glaucoma has no subjective symptoms and it is not possible to regain lost visual fields and visual acuity by treatment, early detection by eye examination and its early treatment are required. Conventional glaucoma diagnosis is often subjective because it strongly depends on the interpretations of color fundus images and optical coherence tomography (OCT) images by medical doctors. In this research, we succeeded to develop a highly accurate machine learning model for glaucoma diagnosis in collaboration with TOPCON corp. and Department of Ophthalmology, Tohoku University. Our model is based on deep learning combined with a random forest algorithm for multi-modal data (Fig. 2) consisting of a color fundus image and four OCT images, and it leads to suppress the number of the training images required to achieve high accuracy. We believe that our model contributes to early detection of glaucoma by representing confidence level of glaucoma.
Automatic Glaucoma Classification: Glaucoma is the result of damage to the optic nerve due to various risk factors such as high intraocular pressure and low blood flow to the retina. Nicolela classification based on optic disc shapes has been widely employed in medical practice of glaucoma diagnosis. It includes important risk factors for glaucoma pathology, and it consists of Focal Ischemia (FI), Myopic (MY), Senile Sclerosis (SS), and General Enlargement (GE) types. Clinical characteristics such as a progression rate of the disease state and lesion locations differ between the types defined in Nicolela classification. Therefore, glaucoma specialists understand the pathophysiology of glaucoma based on this classification and decide the proper treatment plan. Conventional glaucoma diagnosis is often subjective because it strongly depends on the interpretations of color fundus images by medical doctors. In this research, we developed a machine learning model for the Nicolela classification in collaboration with TOPCON corp. and Department of Ophthalmology, Tohoku University. Our model is based on machine learning with a neural network and feature selection technique, with 91 features including ocular parameters (Fig. 3) extracted from images obtained by the fundus examination device (OCT, etc.) of glaucoma patients as the input data. Our results achieved 87.8 % accuracy for glaucoma classification, and we believe it would contribute to objective glaucoma diagnosis by representing confidence level of its classification.
Digital Image Processing
Fig.1 Noise Reduction by Fast Feature-preserving Filter.
Fig.2 Multiresolutional Analysis.

Fig.3 Tone Mapping of HDR Images.
Recent advances in image data acquisition technology increase importance and amount of image data rapidly. The digital data in natural science (obtained by using medical CT and MRI, confocal laser scanning microscopes, telescopes, satellite, etc.) and in industrial engineering (CT, surveillances, etc.) obtained by acquisition devices are stored as digital image data in computer. Also, digital image data had been become popular to non-experts because of digital cameras and mobile/cellular phones equipped with cameras. On the other hand, development speed of image processing algorithms/methods is relatively slower than the rapid advances in acquisition devices. Therefore, we are working on developing the new and efficient algorithms (computational methods) for such acquired images. Especially, our research includes the following noise reduction, feature analysis, and image synthesis.
Noise Reduction: Real-world images do not exist without noise. Noise reduction with preserving image features have been an intensive and important research topic. In this research, we proposed a new fast and accurate approximation method for famous bilateral filtering. We have achieved significant speed-up for time-consuming noise reduction calculations (Fig. 1) by using our method.
Feature Analysis: Analyzing image features is very useful and a fundamental technique in computer vision and pattern recognition. In this research, we are working on developing the new image features which are based on combining multiresolutional analysis (Fig. 2) and digital geometry processing techniques.
HDR Image Synthesis: High dynamic range image (HDRI) which consists of multi-range intensity distribution can be obtained by taking the photographs for the same target with different exposure settings. It includes bright, dark, and in-between them parts of the scene. Recent commercial digital cameras are equipped with such multiple exposure functions, and provide the HDR images. The image synthesis is necessary in order to display such HDR images in standard display devices. In this research, we developed a fast and accurate method to apply the tone mapping operation on the HDR image to obtain the synthesized image (Fig. 3). The method is based on our fast bilateral filter.
Volume Segmentation of Biological Image
Segmentation and visualization of organelle
regions from the live cell images via VCAD.
Recent advances in real-world data acquisition technology include X-Ray CT and MRI made us possible to obtain the detailed image data such as micro-structures of organs, bones, etc. It has became a very important research subject to extract various information from the obtained image data in order to apply sophisticated data analysis techniques e.g. volume and area computations of iso-values in the volume. The pattern recognition is necessary: the computer requires to know which regions are occupied with the target objects before processing the data analysis. We develop a new volume segmentation system based on statistical pattern recognition methods for one of the fundamental technologies developing in the team.
Thickness Analysis of Retinal Layers based on Medical Axis Transforms
High myopia is an eye disease that is difficult to correct eyesight by glasses due to pathological distortion of eyeballs. This distortion makes retina thinner, and it causes evere eye diseases such as glaucoma and retinal detachment. This research developed a method for computing thickness of retinal layers from 3D images acquired from OCT (optical coherency tomography) by using medial axis transforms. Experimental results show that our method can detect thin-parts around distorted regions, or a clue of high myopia. This is useful for early diagnosis of high myopia and other eye diseases.
Comparison of retinal thickness of GCC (Ganglion cell complex). Foggy-like patterns appear around blood tubes where retinas get thinner.
Vectorization and Quality Improvement for Low Resolution and Noisy Images
We propose a algorithmic method to improve the quality of noisy and low resolution images. The improved images are suitable for further image analysis.
In our algorithm, the input raster image (a set of points with intensity) is approximated with a continuous function (vectorization). And then, resampling the scalar field with arbitrary resolution, a high resolution image can be obtained. Converting the raster image into a continuous function provides many good properties including smooth representation and availability of quality differential values. These properties are suitable for image analysis. The output raster image inherits these properties.
The key idea of this algorithm is a good approximation of intensity. We adopt the Partition of Unity (PU) approach which is noise-robust and generates a precise approximation function. This algorithm consists of a set of local computations which provides us the fast and efficient approximations even for a large volumetric image. Unfortunately, the reconstructed image edges may be overly smoothed because of the PU's smooth approximation property. We solved this problem by introducing the new surface-based edge detection and preservation techniques into our algorithm, and achieved the high quality edge-preserving approximations.
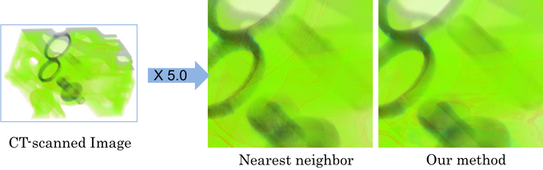
Image Vectorization via Edge-preserving PU.
Digital Geometry Processing

Fig.1 Feature Line Extraction.

Fig.2 Geometric Shape Deformations.
We develop the new efficient algorithms of geometry processing for geometry/shape information obtained from the real-world. The developed algorithms include the following subjects.
Feature Line Extraction: The surface creases (Ridges, Crest Lines) have been widely used in shape recognition and data compression. We re-discover the formula of principal curvatures and Laplace-Beltrami operator of the surface normal, see Fig. 1. By using the formula, a new fast feature line extraction algorithm is proposed. The algorithm can process about 1 million per second to calculate surface differential properties. Here n is the surface unit normal, triangle symbol and inverse triangle symbol are the Laplace-Beltrami operator and surface Gradient operator, respectively.
Geometric Shape Deformations: Shape deformations are very important technique in digital entertainment industry and CAD. We proposed a new thickness-preserving shape deformation method by using the medial axis, see Fig. 2.
Image-based Shape Modeling
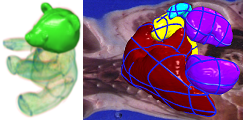
Fig.1 Segmentation via Bilateral H-RBF.

Fig.2 CT-based Flower Modeling
We have been working on integrating digital image and geometry processing techniques.
Interactive Volume Segmentation: Most of the time, a target region for segmentation is not enclosed by distinctive edges (especially 3D images). Expert's knowledge and educated guess are necessary to extract such regions, e.g. biomedical images. In this research, we proposed a novel interactive technique to segment a volumetric region. Our technique is based on a new formulation of Hermit-type RBFs (radial basis functions); Bilateral H-RBFs. Our BH-RBF successfully interpolates a given curve network and passes through image edges (Fig.1).
Flower Modeling from X-ray CT: Modeling flowers in computer is important not only for digital entertainment such as game and cinema, but also for botanical art, digital identification manuals, and physical simulations. Modeling flowers is not easy because of their occluded thin layered surfaces. In this research, we have developed a novel flower shape modeling technique using X-ray CTs of real-world flower samples. Our technique is based on a set of new active contour/surface models which extract flower surfaces from a X-ray CT, interactively (Fig.2).
3D Imaging Device for Detailed Texture Objects

Fig. 1 Insect data generated by our device.
We develop a new 3D imaging device specialized to provide 3D models which consist of geometry information and corresponding photo-realistic textures from real-world small-sized objects.
For small-sized objects (about 3cm to 10 cm), it is difficult to obtain the target geometry and its corresponding textures simultaneously by using conventional commercial imaging devices, because their DOF (depth of field) are not designed to focus on such small-sized objects. Therefore, we develop the imaging device which equipped with the 1mm DOF light source. The device allows us to capture the geometry and its detailed textures of the small-sized objects. Fig. 1 shows the imaging result via our device, where the 3D computer model is generated from a insect specimen. Our device also enables to combine the generated 3D model and motions captured from living insects. Application of the device includes digital encyclopedias of biological and historical objects.
Interactive 3D Web Viewer for Chromatic Volumes

Fig. 1 Interactive 3D Web Viewer
We are developing a new interactive web viewer for 3D chromatic images. The viewer is capable to run interactively on a web browser of standard PC.
While 3D images generated by standard X-ray CT and MRI consist of the transmittance of objects being scanned, 3D internal structure microscopes (3D-ISM) generate chromatic volumes consist of red, green, and blue color channels. Although visualization of former cases is well studied by using volume rendering techniques, the interactive volume rendering with operations (rotation, magnification, slicing, etc.) for the latter is much difficult (especially without high-end graphics cards). We are developing the interactive 3D web viewer specialized to visualize chromatic volumes as Fig 1.
Our viewer includes a stereoscopic visualization function which provides intuitive understanding of 3D volume information. We expect that our viewer contributes to researchers who based on chromatic volumes by assisting to open their data to the public through the Internet.
Local-to-Global Animation Framework for Self-Actuated Flexible Objects
Fig. 1 Tensor Field Editing.
Fig. 2 Animation Result.
We develop a new animation framework for self-actuated flexible objects such as muscular tissues or mollusks.
Animations of flexible objects are difficult to design using traditional skeleton-based or keyframing-based approaches. We then introduce a new local-to-global deformation framework; we synthesize global motions procedurally by integrating local deformations. When the user specifies muscle fiber orientations over the model (fig 1a) and contraction patterns along the fiber orientation (fig1b), our system generates the global motion (fig2). We also present a fast and robust deformation algorithm based on shape-matching dynamics, allowing the user to interactively add external forces on the animated objects.
Endoskeleton Robot Suit: StillSuit
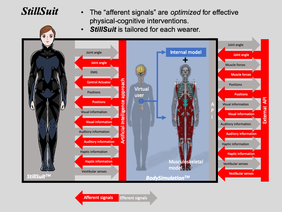
StillSuit Concept ((c)IEEE, LN: 4835600188094, 2020).
In order to solve the near future problems of Japan's aging society, this research project aims to reduce the social burden of medical care and nursing and to secure labor resources. Our approach is based on extending human’s healthy life expectancy by biological human augmentation technology instead of using traditional medical care techniques. We have been developing a robot suit (StillSuit) in collaboration with RIKEN and AIST as an implementation of such human augmentation, in which is an integrated human augmentation of cognition and machine-assisted exercise. Our project also aims to human understanding which includes from genetic information to the mind.
Image Processing for Resonance Bio
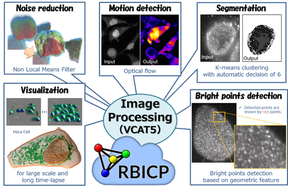
Examples of digital intracellular logistics.
Image processing is one of the most advanced research fields in computer science, and its medical applications such as image diagnosis have been also widely studied. On the other hand, in-vivo image processing on the order of micron, such as intracellular fluorescent dyes and cell populations, is still under development. Thus, the project "Resonance Biology for Innovative Bioimaging" aims to dramatically improve bio-imaging technology through interaction between researchers designing molecules and controlling light, because of increasing demands of solving both quantitative and qualitative problems of bio-images due to imaging technological advances.
In this research, we have developed several useful image processing methods, such as fast filters and deep learning of medical/biology images, for the project. We have also developed a cloud-based image management platform (RBICP), an integrated image processing software (VCAT5) including the proposed methods (and other popular image processing techniques) as plug-in programs, a performance evaluation system for image recognition (Sommelier), and a near infrared imaging system. Our systems have employed for various bio-image analysis. Furthermore, we held three algorithm contests using bio-images and contributed to integrating informatics and biology.
Digital Analysis for IntraCellular Logistics
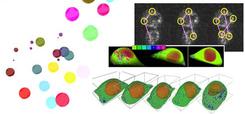
Examples of digital intracellular logistics.
Global mechanism of each membrane traffic such as endocytosis, exocytosis, traffics between organelles, etc. has been recently considered to play a key role for understanding of pathology. Let us call such integrated membrane traffic mechanism by intracellular logistics. Especially it is becoming more and more popular because some of their time-lapse data observation recently became possible by live cell imaging technologies. However, the mainstream in traditional cell biology is microscopic analysis for some particular membrane traffic based on the observed data. Also, these observed data interpretations strongly depend on individual observers.
In order to tackle the above problems, we propose to develop the quantitative digital analysis methods which are less heuristics for intracellular logistics analysis. Developing a unified and standardized approach which is suitable for the most membrane traffics would be the most urgent problem in terms of robust pathological interpretation for the observed data. Therefore, we have started the research of imaging methods and image processing algorithms for standardized quantitative analysis of intracellular logistics data at April, 2008. Our research contributes to investigating intracellular logistics by using our VCAD system which has strong capabilities to process multi-material living objects. We also expect that our research will become very important for fusion of state-of-art cell biology and advanced image processing.
-
Research Contents:Digital Analysis for IntraCellular Logistics (Japanese Only)
Live Cell Modeling
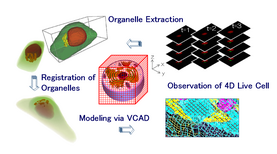
Example of live cell modeling.
The new interrogation and research technologies for understanding of living cells are required in order to integrate the results of traditional and present cell-biology which usually concentrates individual phenomenon deeply, i.e. new tools to discover what the essences of the live (organism) are. Therefore, we are working on constructing the computer models of the living cell for computational analysis to consider the living cell from a global view of the cell-biology. The models are based on the observed data via confocal laser scanning microscopes under standardized and quantitative conditions. The first step of such work has been took in the project "Bio-research infrastructure tools for live cell modeling", a Strategic Programs for R&D (President's Discretionary Fund) of RIKEN. The project consists of 12 laboratories across 6 research centers in RIKEN. We have been developing the new techniques of biological experiments, quantization and acquisition of living cells, 4D live cell imaging, cell image and volume processing, and 4D model construction.