Chemical genetic being conducted in our laboratory is a novel approach to basic biology using specific bioactive molecules as a leverage. To obtain unique chemicals that can achieve the major breakthrough, we always make great efforts to develop assay systems for chemical screening based on each researcher’s idea. For instance, we have developed a screening system to identify inhibitors for human gene products by detecting activity of rescuing the growth defect caused by overexpression of the particular human gene product in the drug-hypersensitive fission yeast cells (Fig. 1). We also developed an in-situ screening system based on digitonin-permeabilized cells (semi-intact cells) to identify a novel SUMOylation inhibitor by visualizing SUMO conjugation activity using GFP-SUMO. Furthermore, we developed reporter assay systems to screen for compounds that induce cell reprogramming and cell differentiation, while altered histone modifications were monitored by a FRET biosensor. Our screening facility established cooperatively with RIKEN Program for Drug Discovery and Medical Technology Platforms (DMP) allows us to perform high-throughput screening.
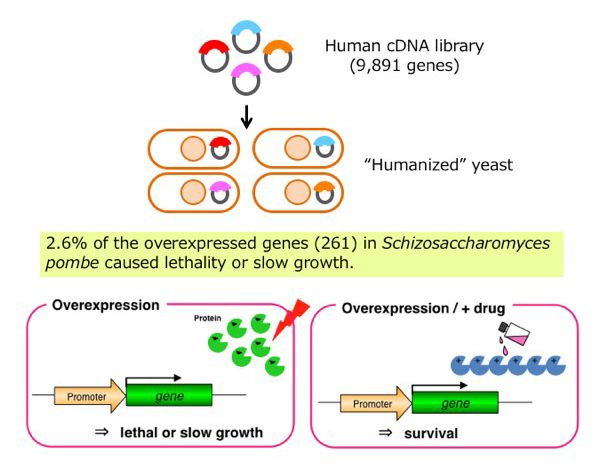
Fig. 1 Identification of human genes that cause growth defect in the fission yeast cells, which can be utilized for developing an inhibitor screening system by means of their overexpression in yeast, called "humanized yeast".
2.Mechanisms of action of bioactive compounds
Unique and surprising biological activity of small molecules attracts interests from a broad range of biological communities. Elucidation of the mechanisms of action of bioactive small molecules is an important start point for not only application of drug development but also creation of the molecular bases for understanding the fundamental life phenomena and discovering new drug targets. By means of chemical genetics that utilizes bioactive natural products showing unique activities (phenotype), we have successfully identified histone deacetylases (HDACs), a protein nuclear export factor (CRM1), a splicing factor (SF3b), etc., as critical regulatory factors for eukaryotic gene expression and new targets for most advanced drug discovery. Unlike genetics that employs mutations, chemical genetics can quickly and reversibly change gene function by chemical methods without causing a permanent damage in the target DNA sequence. In this sense, it is suitable for functional analysis of genes of higher eukaryotes, which are highly complex and redundant. Nevertheless, the successful elucidation of the mechanisms of action of biologically active substances has traditionally required trial and error processes depending on the experience and intuition of a researcher. Therefore, we have been establishing experimental systems to identify drug target molecules logically and systematically by combining post-genomic tools and synthetic chemistry approaches based on cutting edge molecular genetics. Our primary strategies are (1) Chemical genomics and reverse proteomics (Fig. 2) based on the cloning and expression of the whole set of fission yeast ORFs (ORFeome), (2) Drug sensitivity tests of global gene knockdown in the pooled cells after transfection with bar-coded shRNA libraries (3) Identification of human cDNAs encoding drug target proteins using a three-hybrid split luciferase (NanoBiT) luminescence complementation system in cultured mammalian cells. By developing and improving these methodologies, we are making efforts to identify targets for bioactive compounds whose mechanisms of action have not yet been clarified.
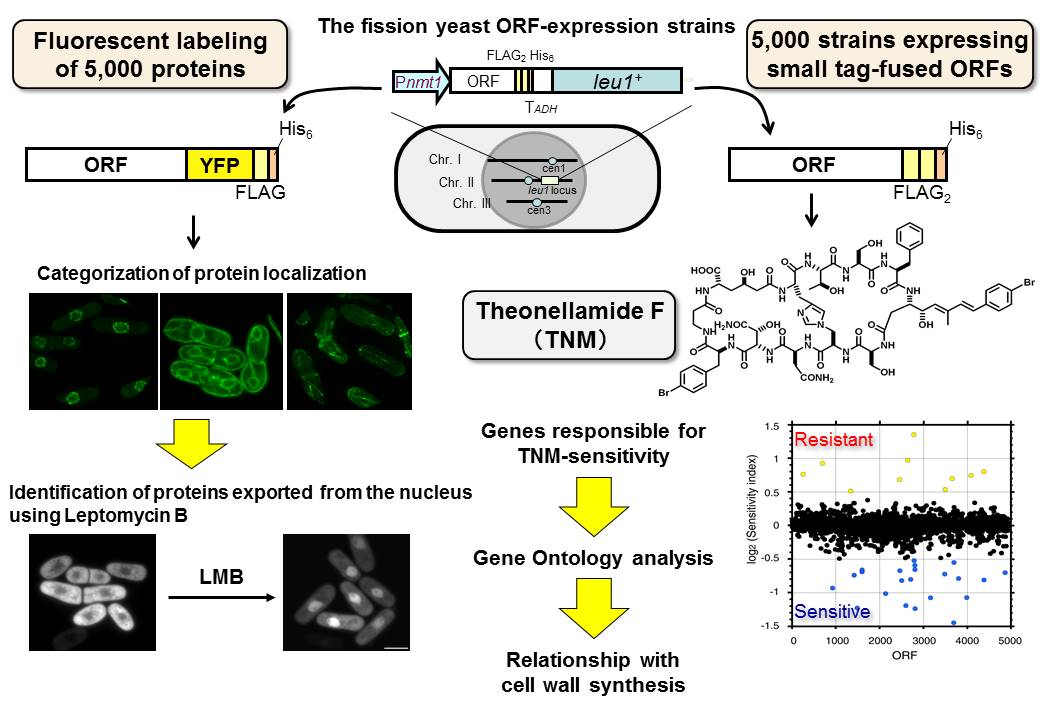
Fig. 2 Chemical genomics using the fission yeast ORFeome.
We have cloned all of 5,000 gene ORFs encoded in the genome of fission yeast. We have constructed several sets of strain collections by inserting the cloned genes into the chromosome one by one. For example, a set of 5,000 strains expressing ORFs fused with YFP (yellow fluorescent protein) can be used for screening for proteins whose subcellular localization is altered by compound treatment. On the other hand, a set of strains expressing ORFs fused with FLAG and His tags can be used for screening for genes whose overexpression alters sensitivity to a compound of interest.
3.Development of live imaging tools to trace the mode of action of bioactive compounds
Epigenetics is defined as the heritable transmission of patterns of gene expression. We focus on the change in epigenetic modifications during various cellular events and are developing live-cell imaging methods to monitor how histone acetylation and methylation are controlled and how small molecules inhibiting the regulating enzymes act in living cells (Fig. 3). We have developed FRET-based fluorescent probes for real-time imaging of histone H4K5/K8, H4K12, and H3K9/K14 acetylation, and succeeded in visualizing the effects of histone deacetylase inhibitors with anti-cancer activity as well as inhibitors of the bromodomain that specifically recognizes and binds acetylated histones in living cells. Furthermore, we are currently constructing imaging probes for histone methylation.
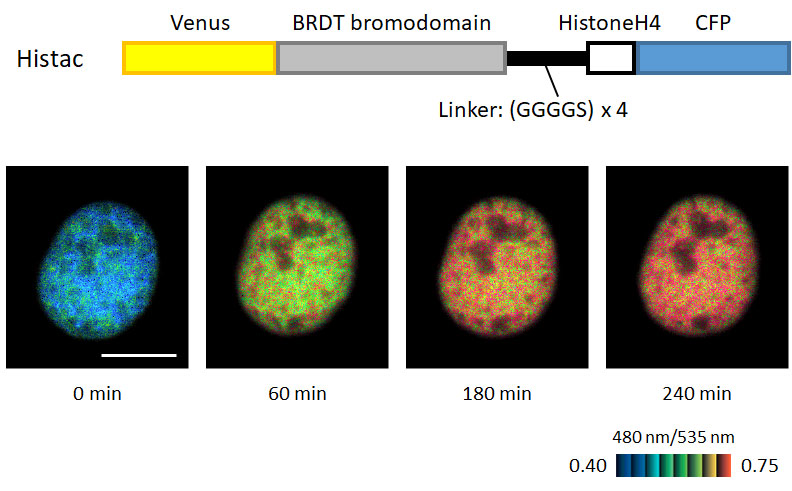
Fig. 3 A fluorescent probe for real-time imaging of histone acetylation in living cells
Construction of a FRET-based acetylation probe (Histac) and real-time imaging of histone acetylation in living cells. Scale bar, 10 µ m.
4.Functional analysis of protein acetylation, and acylation
We have identified the first specific inhibitor of proteins deacetylases (HDACs or KDACs) about 30 years ago. HDAC inhibitors were found to induce strong anticancer activity, some of which have already been used as clinical drugs. However, the detailed mechanism underlying their anticancer activity still remains elusive. This is partially because not only histones, but a variety of other proteins are also deacetylated by HDACs. Therefore, it is necessary to identify biologically important proteins whose function is regulated by acetylation. Indeed, we have so far found that acetylation of alpha-tubulin, a component of microtubules, stabilized the microtubule network, and that acetylation of T-antigen, one of the viral oncoproteins, enhanced its protein stability. Our recent study further revealed cancer-related proteins subject to functional acetylation, including cortactin that regulates cell motility involved in cancer metastasis or invasion. We continue to elucidate the function of acetylation in many regulatory proteins.
Recent studies have shown that some proteins are subject to long-chain fatty-acylation, in addition to acetylation. HDACs have appeared to be responsible for not only deacetylation but also the defatty-acylation. We recently found that SIRT2, an NAD+-dependent deacetylase, acts also as a deacylase for long-chain fatty-acylated proteins. We also identified some novel SIRT2 specific inhibitors, which bind the hydrophobic cavity responsible for the site to recognize the fatty acyl-moiety of substrates. Interestingly, the SIRT2 inhibitors inhibit the deacetylase activity, but not the defatty-acylase activity of SIRT2. During the course of our research to uncover the mechanism by which the SIRT2 inhibitors can selectively inhibit deacetylase but not defatty-acylase, we found the role of intrinsic lipids in controlling the SIRT2 activity. Now we are conducting further analysis aiming at elucidating the mechanism by which sirtuins regulate cellular acetylation and acylation in response to nutrition states.
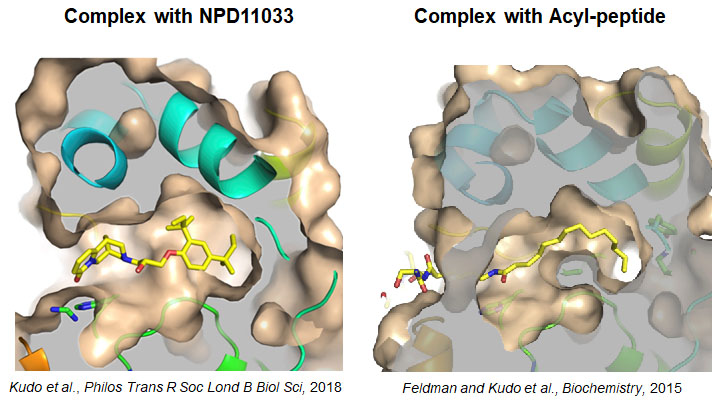
Fig. 4 Crystal structures of SIRT2 in complex with a specific inhibitor NPD11033 (left), and with a substrate, a peptide containing long-chain fatty-acylated lysine (right).
In the inhibitor complex, a hydrophobic pocket is formed upon binding of the inhibitor. The pocket is also formed in the presence of the acylated substrate. These crystal structures suggest that the pocket is for acyl-substrate binding.
5.Mode of action of the splicing inhibitor Spliceostatin A.
The natural product FR901464 was identified as an anticancer agent by the Fujisawa pharmaceutical company, but its mechanism of action remained unknown. Our lab, using FR901464's methylketal derivative, later named Spliceostatin A (SSA), and its biotin conjugate identified the SF3b subcomplex of the U2 snRNP as the molecule's target. SSA prevents the excision of intronic sequences from pre-mRNA. Yet, some improperly spliced transcripts leave the nucleus and become translated into protein, often resulting in the production of truncated polypeptides. Therefore, SSA provides the first potent tool to understand how defective mRNAs can circumvent control mechanisms and escape from the nucleus. It appears highly plausible that intronic sequences hold yet undiscovered information guiding biological processes and mRNA metabolism. Using SSA we are investigating mechanisms of gene expression and their cellular effects. This includes changes to chromatin and also effects on cellular stress responses and translation.
6.Development of systems regulating energy metabolisms in microorganisms and microalgae
The application of chemical biology is not limited to drug discovery. We are also trying to solve global social problems such as carbon neutral energy production using chemical biology. One of such the projects is to apply small molecules for energy production from microorganisms. We are aiming to control productivity of alcohol fermentation in yeasts and lipid synthesis in microalgae by compounds through understanding the energy metabolisms of these microorganisms. For example, we focus on a posttranslational modification called “hypusination” occurring on the specific lysine residue in the translation factor eIF5A, which may be a novel mechanism to activate mitochondrial respiration in response to increased oxygen concentrations. We have revealed that hypusination, a reaction that requires oxygen, is impaired when the ambient oxygen concentration is decreased, leading to reduced mitochondrial function. Indeed, the mutants defective in hypusination-related enzymes showed decreased oxygen consumption, while the alcohol fermentability was increased. We are therefore trying to make eIF5A hypusination controllable with compounds.
To induce oil production in microalgae such as Chlamydomonas, it is necessary to limit the nitrogen content in culture, although an enough amount of the nitrogen source is essential for cell proliferation. To maximize the productivity of oils, the nitrogen source should be supplied until the cells are fully grown and then be removed upon inducing oil production. However, the process to remove the nitrogen source requires a non-negligible amount of energy. Therefore, we are screening for compounds that induce oil production even in the presence of the nitrogen source.
Microorganisms utilize nitrogen and carbon sources in a coordinated manner. We recently found a novel cell-to-cell communication system that regulate nitrogen metabolism, mediated by a secreted small molecule called "nitrogen signaling factor (NSF)" in the fission yeast Schizosaccharomyces pombe. Normally S. pombe does not uptake non-preferred nitrogen sources such as branched-chain amino acids when high-quality nitrogen sources such as ammonium and glutamate are simultaneously present, which are preferentially utilized rather than non-preferred nitrogen sources. However, NSF enables yeast cells to utilize non-preferred nitrogen sources even in the presence of preferred nitrogen sources. Interestingly, once the cells became able to uptake non-preferred nitrogen sources by NSF, the ability was sustained over generations even without supplying NSF. We are currently investigating the intracellular signal transduction system triggered by NSF and a mechanism for keeping the nitrogen metabolic memory.
7.Development of a technique for regulating fungal denitrification
The balance of global nitrogen has been maintained by the microbial processes of the nitrogen cycle consisting of nitrogen fixation, oxidation, and denitrification. However, the invention of the synthetic ammonia process (Haber-Bosch process) in the early twentieth century enabled us to produce huge amounts of nitrogenous fertilizer to be introduced into agricultural lands, which resulted in a dramatic progress in food production, saving people from starvation, and eventually the explosion of the world’s population. At the same time, excess nitrogen in the soil destroyed the natural balance, causing serious global environmental issues such as pollution of water bodies and atmosphere. In particular, a dramatic increase in the nitrous oxide (N2O) concentration in the atmosphere is regarded as one of the most serious environmental risks, because it has global warming potential 300-fold higher than that of carbon dioxide (CO2) and is also recognized as one of the most dominant ozone-depleting substances. Ammonia is converted to nitrate by nitrifying bacteria, and nitrate is not only taken up as nitrogen sources by plants but also used by microbial denitrification called anaerobic respiration (Fig. 5). In bacterial denitrification, the N2O gas is an intermediate to the nitrogen. Recently, denitrification activities were also observed among filamentous fungi inhabiting the aerobic environments. Unlike bacterial denitrification, the end product of fungal denitrification is N2O, because they lack nitrous oxide reductase (N2Or), the enzyme that catalyzes the final step of the denitrification pathway, suggesting that the fungal denitrification is the dominant source of N2O generated in agricultural soils. Therefore, we expect that prevention of fungal denitrification will lead not only to the improvement of nitrogen uptake by plants resulting in the reduction of the amount of nitrogenous fertilizer applied to agricultural soils, but also to the prevention of N2O emissions. Thus, we are challenging the development of eco-sustainable fungal denitrification inhibitors.
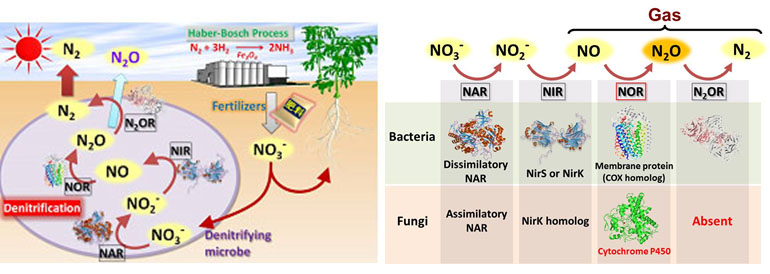
Fig. 5 Development of a technique for preventing greenhouse gas N2O emissions from agricultural soils
We develop compounds that inhibit fungal NIR or NOR, the key enzyme of denitrification pathway, which are expected to lead to the improvement of nitrogen uptake by plants and the reduction in nitrous oxide emissions.
8.Studies on aging control
(1) Analysis of lifespan extension by metabolites
Successful identification of genetic mutations that extend lifespan in model organisms raised the possibility of the discovery of small molecules controlling aging and lifespan in humans. We found that the supplementation of a metabolite in the medium extends chronological lifespan of fission yeast. Metabolome and transcriptome analysis suggest that it increases the intracellular glutathione level and alters gene expression of many genes. Genetic screening to identify genes involved in the process of lifespan extension is in progress.
(2) Screening for chemical compounds that improve cellular energy metabolism
Growth of cells derived from cancers or patients suffering from mitochondrial disease depends on glycolysis. We searched for chemical compounds capable of shifting metabolism from glycolysis to mitochondrial respiration and identified an active substance. Remarkably, this compound restored neuronal differentiation in iPS cells derived from a mitochondrial disease patient whose iPS cells were previously unable to differentiate into neural cells. Detailed analyses on the mechanism of action of this compound are underway.
(3) Harnessing reactive oxygen species to shift human mtDNA from heteroplasmy to homoplasmy
Genetic mutations occur in mitochondrial DNA (mtDNA) at a frequency much higher than in nuclear genomic DNA. MtDNA mutations accumulate with age in certain human tissues leading to a high proportion of defective mtDNA copies within cells, causing respiratory defects. Similarly, fusing yeast cells harboring wild-type mtDNA with those harboring mutant mtDNA creates an artificial mtDNA mixture, termed heteroplasmy. Using this system, we found that recombination-driven mtDNA replication depends on reactive oxygen species and that one mtDNA allele, either wild-type or mutant but not both, acquires a replication advantage leading to the homoplasmy formation. We further revealed that subjecting heteroplasmic cells derived from patients of mitochondrial disease to an optimal dose of oxidative stress promotes the homoplasmy formation. These results suggest that reactive oxygen species activate the recombination-dependent mtDNA replication mode, which promotes mtDNA segregation. We are currently applying these discoveries for potential therapy of mitochondrial genetic disorders by designing a novel treatment means.
 RIKEN Center for Sustainable Resource Science (CSRS) Chemical Genomics Research Group Japanese
RIKEN Center for Sustainable Resource Science (CSRS) Chemical Genomics Research Group Japanese